Our research group proudly maintains FoodMicrobionet, a metataxonomic database on the composition of bacterial and fungal communities of foods and food environments. The structure of the database and its relationships with external databases is shown below:
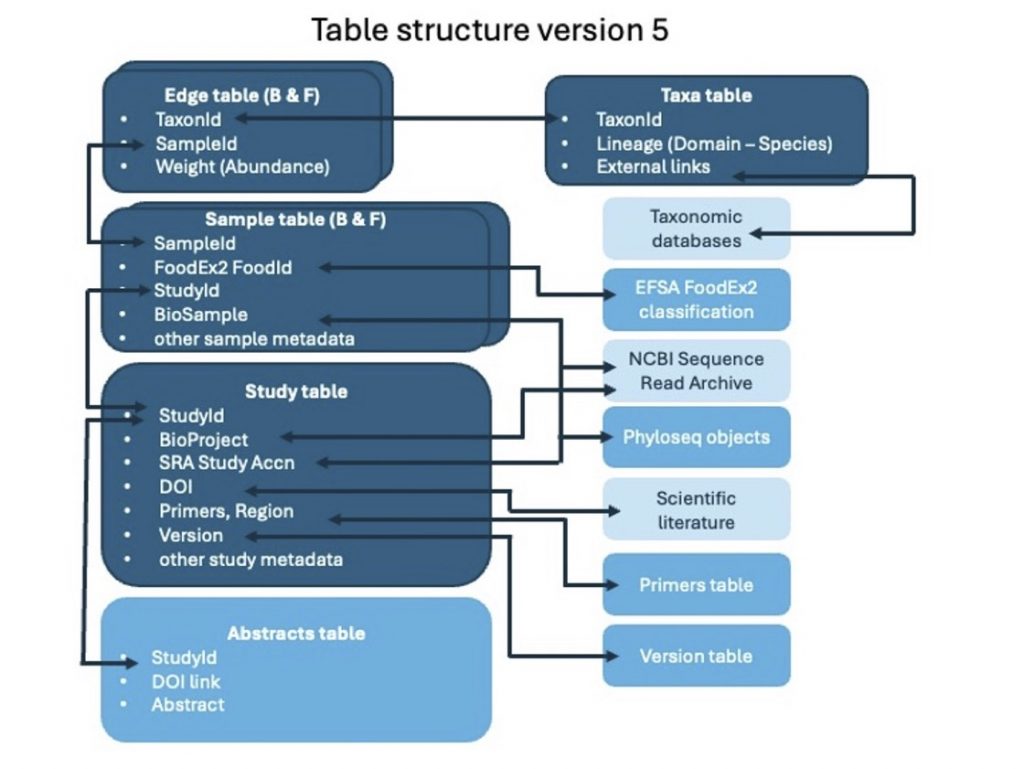
Current version (5.1.2) includes 302 studies with 17,174 samples for bacteria and 3,175 samples for fungi: it is arguably the largest and best annotated database on the structure of food bacterial communities.
The database and scripts for FoodMicrobionet are publicly available on GitHub. In addition, the interactive Shiny app (ShinyFMBN) which allows a simplified access to the database (for searching, selecting studies and samples, expiring data and visualising and exporting graphs is also available on Mendeley data (DOI 10.17632/8fwwjpm79y.7). Finally, processed studies using to feed FoodMicrobionet are available on Zenodo (DOI 10.5281/zenodo.6954040) as R lists.
If you want to keep up to date with latest developments visit the GitHub repository or search this site using FMBN as a tag.
If you want to learn more about FoodMicrobionet and how it can be used for meta studies read our publications:
- Parente, E., Ricciardi, A., 2024. A comprehensive view of food microbiota: introducing FoodMicrobionet v5. Foods 13, 1689. https://doi.org/10.3390/foods1311168
- Parente, E., Zotta, T., Giavalisco, M., Ricciardi, A., 2023. Metataxonomic insights in the distribution of Lactobacillaceae in foods and food environments. Int J Food Microbiol 110124. https://doi.org/10.1016/j.ijfoodmicro.2023.110124
- Parente, E., Zotta, T., Ricciardi, A., 2022. FoodMicrobionet v4: A large, integrated, open and transparent database for food bacterial communities. Int J Food Microbiol 372, 109696. https://doi.org/10.1016/j.ijfoodmicro.2022.109696
- Parente, E., Zotta, T., Ricciardi, A., 2022. A review of methods for the inference and experimental confirmation of microbial association networks in cheese. Int J Food Microbiol 368, 109618. https://doi.org/10.1016/j.ijfoodmicro.2022.109618
- Parente, E., Ricciardi, A., Zotta, T., 2020. The microbiota of dairy milk: A review. Int Dairy J 107, 104714. https://doi.org/10.1016/j.idairyj.2020.104714
- Parente, E., De Filippis, F., Ercolini, D., Ricciardi, A., Zotta, T., 2019. Advancing integration of data on food microbiome studies: FoodMicrobionet 3.1, a major upgrade of the FoodMicrobionet database. International Journal of Food Microbiology 305, 108249. https://doi.org/10.1016/j.ijfoodmicro.2019.108249
- De Filippis, F., Parente, E., Zotta, T., Ercolini, D., 2018. A comparison of bioinformatic approaches for 16S rRNA gene profiling of food bacterial microbiota. International Journal of Food Microbiology 265, 9–17. https://doi.org/10.1016/j.ijfoodmicro.2017.10.028
- Parente, E., Zotta, T., Faust, K., De Filippis, F., Ercolini, D., 2018. Structure of association networks in food bacterial communities. Food microbiology 73, 49–60. https://doi.org/10.1016/j.fm.2017.12.010
- Parente, E., Cocolin, L., De Filippis, F., Zotta, T., Ferrocino, I., O’Sullivan, O., Neviani, E., De Angelis, M., Cotter, P.D., Ercolini, D., 2016. FoodMicrobionet: A database for the visualisation and exploration of food bacterial communities based on network analysis. International Journal of Food Microbiology 219, 28–37. https://doi.org/10.1016/j.ijfoodmicro.2015.12.001